How sex chromosomes break up to get together
Sex chromosomes must exchange genetic information at a short region during meiotic cell division. Molecular factors have now been found that alter sex-chromosome structure and enhance this exchange process.
by Ericka Humphrey & Francesca ColeSperm and eggs are produced through a specialized type of cell division called meiosis, which ensures that the cells have only half the usual number of chromosomes — one copy of each chromosome instead of two in mammals. During meiosis, each set of parental chromosomes exchanges DNA sequences at equivalent (homologous) regions along their length. This exchange of information — a process called recombination — is induced by DNA double-strand breaks that occur about every 10 megabases [1]. However, X and Y chromosomes share only a short (about 700 kilobase) homologous region called the pseudoautosomal region (PAR); double-strand breaks must therefore occur [2] much more frequently in this region to ensure proper recombination between X and Y chromosomes. Writing in Nature, Acquaviva et al. [3] show how a DNA element provides a platform for enhancing the frequency of double-strand breaks in the PAR.
The early part of meiosis consists of several steps. Each chromosome is duplicated, forming two identical copies, known as sister chromatids, that are connected to each other by cohesin protein complexes. Double-strand breaks are then formed along the chromatids by the protein SPO11, with the help of its accessory factors. The 3’ ends of the breaks search for homologous sequences — this leads to pairing and alignment of homologous chromosomes (such as the X and Y, or both copies of any non-sex chromosome) along their lengths, in a process called synapsis. Finally, during recombination, chromatid arms are exchanged between the two homologous chromosomes. This last step is essential for the accurate segregation of each pair of homologous chromosomes into separate daughter cells. Insufficient recombination at the PAR is a major cause of developmental disability and infertility [4]. Indeed, the sex chromosomes are the most frequently mis-segregated chromosomes in sperm [4].
The frequency of meiotic double-strand breaks correlates with the number of chromatid structures called ‘loop–axis units’. DNA loops emanate from a protein-rich DNA axis that makes up the backbone of the chromatid [5] (Fig. 1). Chromatid regions that have longer axes and shorter loops generally have more double-strand breaks than do those with shorter axes and longer loops. The PAR has a relatively long axis for the length of its genomic sequence, suggesting that chromosome structure might promote the high frequency of breaks needed to ensure PAR recombination [1].
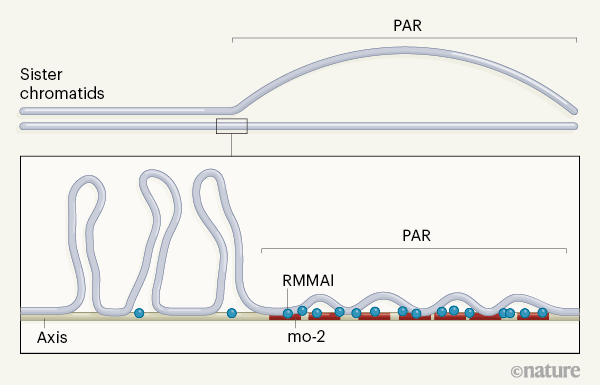
How might this PAR-specific chromosome structure be formed and regulated? To answer this question, Acquaviva et al. analysed mouse spermatocytes (meiotic precursors of sperm). They found that the PAR is restructured before synapsis, both to elongate the axes of aligned sister chromatids and to separate them from one another at the PAR (Fig. 1).
The authors showed that the separated axes are strongly bound by five proteins that promote double-strand-break formation (REC114, MEI4, MEI1, ANKRD31 and IHO1; collectively dubbed RMMAI). These factors were already known to accumulate elsewhere in the nucleus [6] [,] [7]; Acquaviva and colleagues found that this accumulation occurs at regions of repetitive DNA sequences called mo-2 minisatellites. The researchers propose that mo-2 minisatellites could act to bind proteins and other molecules involved in chromosome restructuring and double-strand-break formation, including RMMAI proteins.
To test this model, the authors compared a mouse strain that naturally has few mo-2 minisatellites with a standard laboratory strain, which has many more. REC114 accumulation was lower in the ‘low’ mo-2 strain than in the ‘high’ mo-2 animals. The group then analysed PAR structure in spermatocytes from offspring of crosses between the low- and high-mo-2 strains. The sex chromosome derived from the high-mo-2 parent always showed high levels of RMMAI enrichment and axis remodelling at the PAR, regardless of whether it was X or Y; by contrast, the PAR derived from the low-mo-2 parent did not. Moreover, Acquaviva and colleagues showed that mo-2 arrays, along with sufficient accumulation of RMMAI, can lead to structural remodelling of non-sex chromosomes — at the end of chromosome 9, for instance.
Does the accumulation of RMMAI and structural remodelling at mo-2 arrays actually cause an increased frequency of double-strand- breaks? The protein RPA2 is concentrated at sites at which recombination is under way — Acquaviva et al. therefore analysed these RPA2 foci as a proxy for double-strand breaks. RPA2 associated highly with mo-2 arrays. Indeed, when synapsis was nearly complete, 70% of mo-2 sites had associated RPA2 foci. Further supporting the idea, the group showed that the overlap between RPA2 foci and mo-2 arrays was significantly lower in mice lacking the RMMAI component ANKRD31, and previous work has shown that X and Y chromosomes do not properly pair up in these animals [6] [,] [7].
RMMAI accumulation therefore seems to be key for promoting double-strand breaks and recombination at the PAR. By contrast, a mechanism involving the protein PRDM9 is known [8] [,] [9] to control the location of most double-strand breaks outside the PAR. The authors found that the frequency of breaks was higher than average at mo-2 arrays on non-sex chromosomes, and showed that these breaks form independently of PRDM9. Thus, mo-2 arrays define chromosomal regions in which PRDM9- independent mechanisms of break formation can occur, with RMMAI accumulation crucial to this process.
Both double-strand breaks and synapsis occur later at mo-2 arrays than at other chromosomal regions in spermatocytes. The double-strand-break machinery is suppressed once chromosomes have fully synapsed [1] [,] [10]. The authors therefore hypothesized that delayed synapsis might underlie the increased frequency of double-strand breaks at mo-2 arrays. To test this, they investigated chromosome structure and break frequency in the meiotic precursors of eggs, oocytes. The X chromosomes of oocytes do have mo-2-rich PAR regions in which RMMAI accumulates, but they do not rely on these regions for synapsis, because the two X chromosomes share homology along their whole lengths. X chromosomes therefore synapse with the same efficiency as do non-sex chromosomes.
Consistent with their model, Acquaviva et al. showed that levels of double-strand breaks are not as high in the PAR of oocytes as in that of spermatocytes. However, the group could trigger high-level break formation on mo-2 arrays by delaying synapsis. Normally, oocytes progress more rapidly through early meiosis than do spermatocytes. This finding therefore suggests that the duration of early meiosis might be differentially regulated to meet the different biological imperatives of sperm and eggs — with a longer time frame allowing vulnerable regions such as the PAR to accumulate sufficient double-strand breaks.
Why the axes of sister chromatids become separated at the PAR, and whether this separation promotes break formation and recombination, have yet to be determined. As a consequence of axis separation, the sister chromatids of spermatocyte sex chromosomes are farther apart than are those of other chromosomes. The authors suggest that axis separation might suppress ineffectual intersister recombination in favour of homologous recombination between chromosomes. In addition, the group posits that chromosome restructuring is linked with the accumulation of a cohesin protein that they see at the tip of the PAR. Cohesin accumulation in this region might increase cohesion between sister chromatids, which in turn would help to stabilize the association between homologous chromosomes. An alternative model is that axis separation might provide more ‘real estate’ on which factors promoting double-strand breaks could assemble at the PAR because the proteins can accumulate between the separated axes. These non-exclusive models provide intriguing fodder for future research.
How double-strand breaks are formed at the PAR has been a conundrum for the field. Acquaviva and colleagues’ model paints an elegant picture of how a genetic element, chromosome structure and the timing of meiosis interact to ensure proper recombination.
doi: 10.1038/d41586-020-01483-6
References
1. Kauppi, L. et al. Genes Dev. 27, 873–886 (2013).
2. Kauppi, L. et al. Science 331, 916–920 (2011).
3. Acquaviva, L. et al. Nature https://doi.org/10.1038/s41586-020-2327-4 (2020).
4. Templado, C., Uroz, L. & Estop, A. Mol. Hum. Reprod. 19, 634–643 (2013).
5. Zickler, D. & Kleckner, N. Annu. Rev. Genet. 33, 603–754 (1999).
6. Boekhout, M. et al. Mol. Cell 74, 1053–1068 (2019).
7. Papanikos, F. et al. Mol. Cell 74, 1069–1085 (2019).
8. Baudat, F., Imai, Y. & de Massy, B. Nature Rev. Genet. 14, 794–806 (2013).
9. Brick, K., Smagulova, F., Khil, P., Camerini-Otero, R. D. & Petukhova, G. V. Nature 485, 642–645 (2012).
10. Thacker, D., Mohibullah, N., Zhu, X. & Keeney, S. Nature 510, 241–246 (2014).