Coronavirus Testing: CRISPR Technology Set To Streamline Viral Testing
by Dan MaloneyIf we could run back 2020 to its beginning and get a do-over, chances are pretty good that we’d do a lot of things differently. There’s a ton of blame to go around on COVID-19, but it’s safe to say that one of the biggest failures of this whole episode has been the lack of cheap, quick, accurate testing for SARS-CoV-2, the virus behind the current pandemic. It’s not for lack of information; after all, Chinese scientists published the sequence of the viral genome very early in the pandemic, and researchers the world over did the same for all the information they gleaned from the virus as it rampaged around the planet.
But leveraging that information into usable diagnostics has been anything but a smooth process. Initially, the only method of detecting the virus was with reverse transcriptase-polymerase chain reaction (RT-PCR) tests, a fussy process that requires trained technicians and a well-equipped lab, takes days to weeks to return results, and can only tell if the patient has a current infection. Antibody testing has the potential for a quick and easy, no-lab-required test, but can only be used to see if a patient has had an infection at some time in the past.
What’s needed as the COVID-19 crisis continues is a test with the specificity and sensitivity of PCR combined with the rapidity and simplicity of an antibody test. That’s where a new assay, based on the latest in molecular biology methods and dubbed “STOPCovid” comes in, and it could play a major role in diagnostics now and in the future.
Bacterial Immunity
To the extent that CRISPR has entered the popular lexicon, it’s understood to be a new and powerful technique for editing the genome of organisms, potentially as a way to treat genetic disorders. That’s certainly a big part of the story, and one that will rightly win someone a Nobel prize within the next 15 years or so, but it doesn’t tell the whole story. The roots of CRISPR gene editing methods lie in the bacterial world, and how these single-cell organisms evolved a sophisticated immune system that bears a strong resemblance to the human immune system.
With all the stress we put on avoiding, detecting, and treating viral infections, it might be surprising to some people that there are thousands of viruses that have evolved to attack bacteria. It is estimated that these bacteriophages, or phages for short, outnumber their bacterial hosts by a factor of ten, meaning that Earth has something like 1030 phage particles, a number that vastly eclipses any organism’s population. And just like any other virus, like influenza or SARS-CoV-2, a phage’s business is to find a suitable host, inject its genetic material, and take over the cell’s machinery to make more of itself, generally destroying the host in the process.
So it stands to reason that bacteria would have evolved mechanisms to detect and evade viral infections. That’s where clustered regularly interspaced short palindromic repeats, or CRISPR, comes into play. CRISPR sequences are essentially a collection of DNA fragments that have been snipped from the genetic material of invading phages. These short sequences, called spacers, are added to the bacterial genome by a set of enzymes known as CRISPER associate (Cas) proteins.
The spacers are the key to bacterial immunity. Along with some of the adjacent repeating sequences in the CRISPR segment, the spacers are transcribed into complementary RNA fragments called crRNA, or CRISPR-RNA. crRNAs bind to proteins like Cas9, a CRISPER-associated enzyme that can both unwind DNA and cut it. The section of the crRNA from the spacer allows it to bind to the phage DNA that the spacer originally came from, which signals Cas9 to snip the invading phage DNA. No phage DNA, no infection, and having the spacer integrated into the bacterium’s genome means that it and its descendants have a memory of that particular phage.
The discovery of the importance of CRISPR in bacterial immunity naturally led to genetic engineering methods based on the CRISPR-Cas enzymology that go far beyond what can be accomplished with earlier molecular biology techniques, which are based mainly on restriction endonucleases. These enzymes, also part of the bacteria immune system, evolved to cut DNA at short, specific base-pair sequences; there are hundreds to choose from, but you’re stuck making cuts at naturally occurring recognition sequences, or using other genetic engineering techniques to insert the sequences where you want them. CRISPR-Cas allows you to dictate exactly where the cuts will occur, regardless of the sequence.
Collateral Damage
The incredible usefulness of CRISPR techniques has led to the discovery of more Cas enzymes, each with different properties. One such enzyme, Cas13, has very interesting and useful properties. Like Cas9, Cas13 uses a crRNA transcribed from the CRISPR region as a template to recognize invading phages. But rather than binding to and subsequently cleaving DNA, Cas13 targets phage RNA, cleaving it at the sequence specified by the spacer in the rRNA. Once that happens, Cas13 gets a bit sloppy, cleaving any RNA fragment within reach, regardless of sequence. It’s a little like cutting your account number from a bank statement, only to then take all those tiny pieces of paper and put them through a shredder, just to be sure.
It’s this nonspecific collateral damage to RNA by Cas13 that lead to the development of a CRISPR method dubbed SHERLOCK, for Specific High-Sensitivity Enzymatic Reporter Unlocking. Developed in 2017 in Feng Zhang’s lab at the Broad Institute of MIT, SHERLOCK uses tiny reporter fragments of RNA that have been tagged on both ends with different markers. After an initial RNA amplification step, Cas13 finds and cleaves its target RNA. Once activated, it keeps chopping up any RNA it can find, including the reporter RNA fragments added to the reaction. The two markers on the reporter are separated from each other by this collateral cleavage only if the target sequence is present, allowing SHERLOCK to detect viral infections with extreme specificity and exquisite sensitivity; in initial diagnostic tests, viral DNA from patients with Zika virus infections could be detected down into the attomolar range, or about 2,000 copies of the virus per milliliter of sample.
For all its utility, though, the SHERLOCK procedure is still a fussy lab process. Like the reverse transcriptase-polymerase chain reaction process that’s currently the gold standard for COVID-19 testing, a SHERLOCK-based diagnostic test would require trained technicians and a well-equipped lab to perform, the potential for cross-contamination as the multiple reaction tubes are accessed, and would still be subject to delays due to shipping samples and returning the results.
To rectify these problems, the Zhang lab came up with a far simpler version of the SHERLOCK assay called STOPCovid, for SHERLOCK Testing in One Pot. They optimized the RNA amplification and Cas19 recognition and cleavage steps to run together in one buffer at a single temperature, reducing the number of manipulations needed for each sample tested. This greatly reduces the risk of lab mistakes, and makes the process simple enough to perform in even poorly equipped labs.
Lab On A Stick
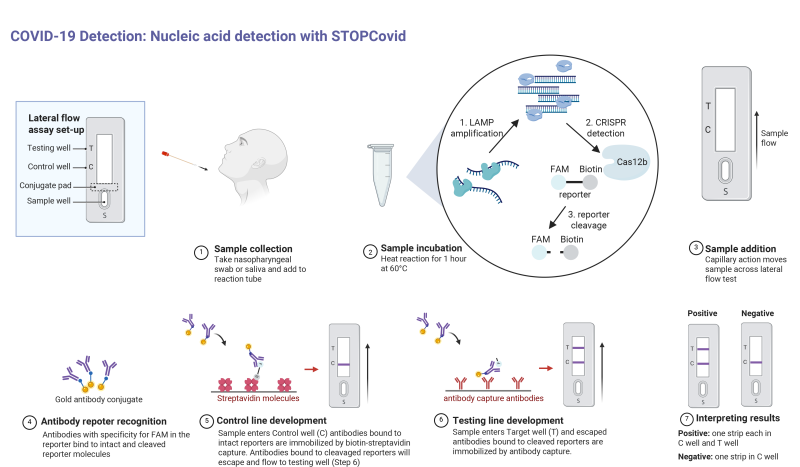
To streamline the process even further, the RNA reporter fragments were re-engineered to allow detection via antibodies. This may seem like a step in the wrong direction, since as we discussed previously, antibody testing for COVID-19 infection can only detect whether a patient has created antibodies to the SARS-CoV-2 virus, and due to the time it takes to do that is more of a lagging indicator of infection. But in the STOPCovid assay, antibodies are used not to detect viral proteins, but as a way to separate the cleaved RNA markers, which are tagged with two different proteins. This opens up the possibility of using a lateral-flow assay, where capillary action pulls reaction solution past strips that have antibodies to the tag proteins applied to them. That will allow health care providers to use a test strip similar to a pregnancy test to read results from the STOPCovid assay, turning the whole process into a point-of-care test.
As with anything in the medical field, there’s a long road between the lab and the clinic. Trials must be conducted, intellectual property disputes must be settled, and manufacturers must be lined up. In the meantime, though, Dr. Zhang and his colleagues aren’t waiting. They’ve made the full STOPCovid protocol available to anyone, with all the details on how to run the reaction and interpret the results. They make it very clear that this is not intended for clinical use at this time, but the mere fact that they’re willing to put the information out there and worry about the details later is pretty encouraging.
With the ability to detect a current infection similar through nucleic acid analysis combined with the ease-of-use of a lateral-flow antibody test, STOPCovid could be a real revolution in viral testing, not only for COVID-19 but for viral infections yet to come.