RDE-3 found to add pUG tails to targets of RNA interference and to transposon RNAs
by Bob Yirka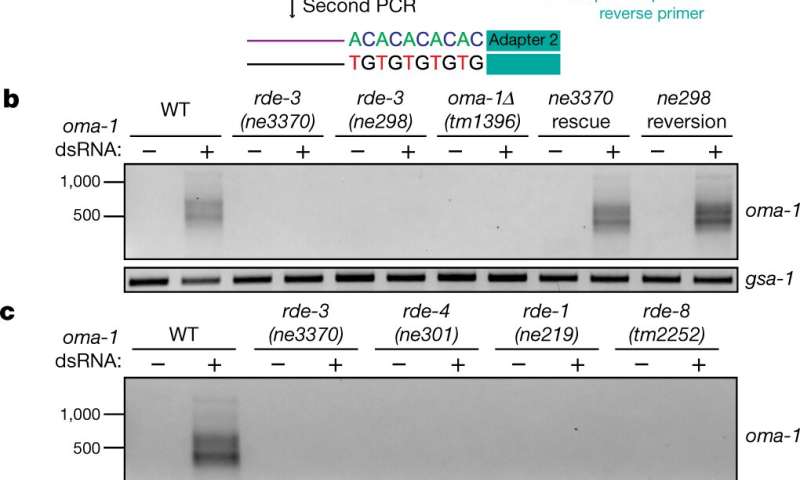
A team of researchers from Harvard Medical School, Nanjing Agricultural University and the University of Wisconsin has found that the protein RDE-3 in nematode worms adds pUG tails to targets of RNA interference and transposon RNAs. In their paper published in the journal Nature, the group describe their findings and the ways their work could impact the study of gene silencing through generations. Kailee Reed and Taiowa Montgomery with Colorado State University have published a News and Views piece outlining the work by the team in China and suggest that the team's finding could pave the way for research into whether poly(UG)-tailed RNAs serve the same role in other species.
As Reed and Montgomery note, small RNAs were discovered in nematode worms almost 30 years ago. And since that time, they have been found to be involved in a surprising number of biological processes. It has also been discovered that small RNAs can be transmitted from one generation to the next, revealing a non-DNA-based form of heritable genesilencing. But until now, this mechanism was carried out was not known. In this new effort, the researchers have found that it is through pUG tails (an abbreviation of poly(UG) tails, via a process called pUGylation.
Prior work had shown that small RNA initiates mRNA silencing by attaching to a Argonaute/Piwi protein and then acting as a guide to control mRNA cleavage. In this new work, the researchers found that the cleaved mRNA is bound by a RDE-3 enzyme. They also found that the RDE-3 enzyme added a tail to the mRNA, which was made up of uridine and guanosine nucleotides. Such pUG tails served as a template for the RNA polymerase enzyme—and it in turn, synthesized secondary small RNAs. And those secondary RNAs, the researchers suggest, maintain mRNA silencing by serving in the same role as initiating small RNA. They further suggest that cycles of truncation and synthesis are the drivers of transgenerational gene silencing. In practical terms, they suggest this cycle is a way for nematode worms to inoculate their offspring against parasites.
More information: Aditi Shukla et al. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance, Nature (2020). DOI: 10.1038/s41586-020-2323-8
Journal information: Nature
© 2020 Science X Network